

netconv2NX() to make the conversion to NX easier.
See NX.
pyNetConv is a Python library created to help the conversion of some
network file formats.
Since version 0.6, pyNetConv is capable to read/write (and convert from/to)
the following file formats:
And from version 0.8 it is able to import tab-separated values exported by Affymetrix Micro Array Suite 5.0 (MAS5) and export this data as Cytoscape node attributes or expression data (.pval file).
pyNetConv is a library to create a software that is able to read/write
networks in the formats supported by pyNetConv. If all you want is convert the
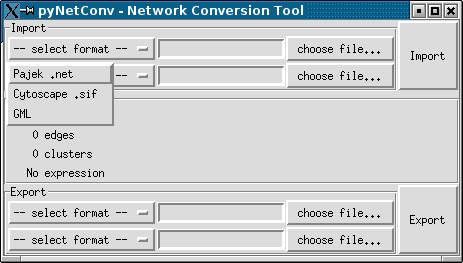
files without the need create a program, then the Graphical user interface is
for you. See the screenshots session to see how the GUI looks
like.
The motivation for creating this tool was that there are many good graph visualization/layout tools available (each one with strengths in some areas) but sometimes they have incompatible file formats (this is true for Pajek and Cytoscape). Some tools can read/write GML files, while other not.
So with pyNetConv we plan to fill this gap allowing the user to exchange
files between this tools and use the best features of each tool to generate
the final graph.
pyNetConv is developed as a library, so one can write his own code and use
pyNetConv to import/export the network in the formats supported.
Example:
n = Network() importPajek(n, 'network.net') importPajekClu(n, 'network.clu') n.cluster2Attributes() n.pajek2pos(1000) exportCyAttributes(n, 'network.cluster') exportGML(n, 'network.gml') # network + cluster information
If you want to import a GML, Pajek or SIF file to use with NetworkX, you can do the following:
import netconv import networkx as NX n = netconv.Network() netconv.importPajek(n, 'network.net') G = NX.Graph() netconv.netconv2NX(n,G) # from here, work with G
Documentation is on plans. For now you can take a look at the pyNetConv API for some documentation.
 |
 |
 |
pyNetConv to
communicate directly with Cytoscape (with this feature, pyNetConv will be able to
modify the network loaded in Cytoscape)
Only Python is required to run pyNetConv. You can get it from the Python web site.
Download the files from the project page.
After unpacking the file, execute:
python setup.py install
in the package new created folder to install it.
To use the graphical interface run the file netconv-gui.py.
For Windows, you can download the Win32 installer (with extension .exe). If you want the documentation of the API, you should also download the source code (it is not included in the Win32 installer).
Moved to a separate file
pyNetConv divides all the positions by the biggest
value, generating values between 0 and 1, but for Pajek to GML, there is no
easy way to guess the value to convert (it depends on many factors, as the
number of nodes, their positions, etc), so the user should inform a value
to multiply all the coordinates in Pajek to put them in the GML file. The
user should find out the one that fits best to his/her network. A good
start is 1000.
netconv-gui.py installed?C:\Python24\Scripts\
| Author: | Márcio Rosa da Silva |
| <mrs (a) gbf de> | |
| http://www.gbf.de/SystemsBiology |
 |
|